A novel mechanism of gastric cancer metastasis was identified by Tianhua ZHOU research group
Gastric cancer is one of the most common and lethal malignancies worldwide1, 2. It is currently both the second most common tumor and leading cause of cancer-related death in China3. The overall 5-year survival rate of gastric cancer patients is about 25%4. Approximately 40% of patients with gastric cancer present with metastases and only about 5% of these patients survive for 5 years 4, 5. Due to the lack of effective therapy, successful clinical management of metastatic gastric cancer remains a major challenge. Therefore, there is an urgent need to learn more about the mechanisms of gastric cancer metastasis, and to develop new therapeutic strategies.
Recently, Tianhua Zhou’s lab from Zhejiang University and Yibin Kang’s lab from Princeton University and published their collaborative work on Gastroenterology (IF 20.7). The article title is “Long Non-coding RNA GMAN, Upregulated in Gastric Cancer Tissues, is Associated with Metastases in Patients and Promotes Translation of Ephrin A1 by Competitively Binding GMAN-AS”. In this article, the researchers have identified a long non-coding RNA (lncRNA) which is highly expressed in gastric cancer tissues and correlates with metastasis and poor prognosis. Therefore, they termed this lncRNA as GMAN (gastric cancer metastasis associated long non-coding RNA). Further studies showed that GMAN is a sense lncRNA which promotes ephrin A1 mRNA translation by competitively binding to the antisense GMAN RNA (GMAN-AS). These data suggest a novel triple RNA competition model for translational regulation, and revealing a novel type of genetic information transmission mediated by sense lncRNA. Moreover, this work identifies an important role of GMAN/GMAN-AS/ephrin A1 axis in gastric cancer metastasis, and suggests a potential new therapeutic approach for gastric cancer metastasis.
Recent transcriptome studies have shown that over 70% of the human genome is transcribed to RNAs, a majority of which are non-coding transcripts (ncRNA)6-8. It can be divided into small non-coding RNA (sncRNA) and lncRNA according to the molecular size of ncRNA. LncRNAs are loosely defined as RNAs that exceed 200 nucleotides in length and exhibit no apparent coding capacity9-11. Intriguingly, lncRNAs play important and versatile roles in gene regulation including epigenetic regulation, transcriptional regulation and post-transcriptional regulation by acting as “guide”, “decoy”, “scaffold” or “signal” 12-15. Aberrant lncRNA expression has been reported to be correlated with various diseases12-15. However, it is not clear how lncRNAs affect gastric tumor development and progression.
In this study, the researchers have identified a previously uncharacterized lncRNA GMAN (gastric cancer metastasis associated long non-coding RNA) which is frequently upregulated in gastric cancer tissues and significantly correlated with metastasis and poor prognosis. To further determine the role of GMAN in gastric cancer progression, GMAN was knocked down or knocked out using small interfering or hairpin RNAs or CRISPR/Cas9 vectors, and it was overexpressed from transfected plasmids in gastric cancer cells. The results showed that GMAN could significantly promote gastric cancer cell invasion and metastasis. Rapid amplification of complementary DNA ends and bioinformatics analyses revealed that GMAN is an 855-nucleotide (nt) sense transcript, which spanned from intron 2 to intron 3 of the ephrin A1 gene. Furthermore, GMAN regulates gastric cancer metastasis by modulating ephrin A1 mRNA translation.
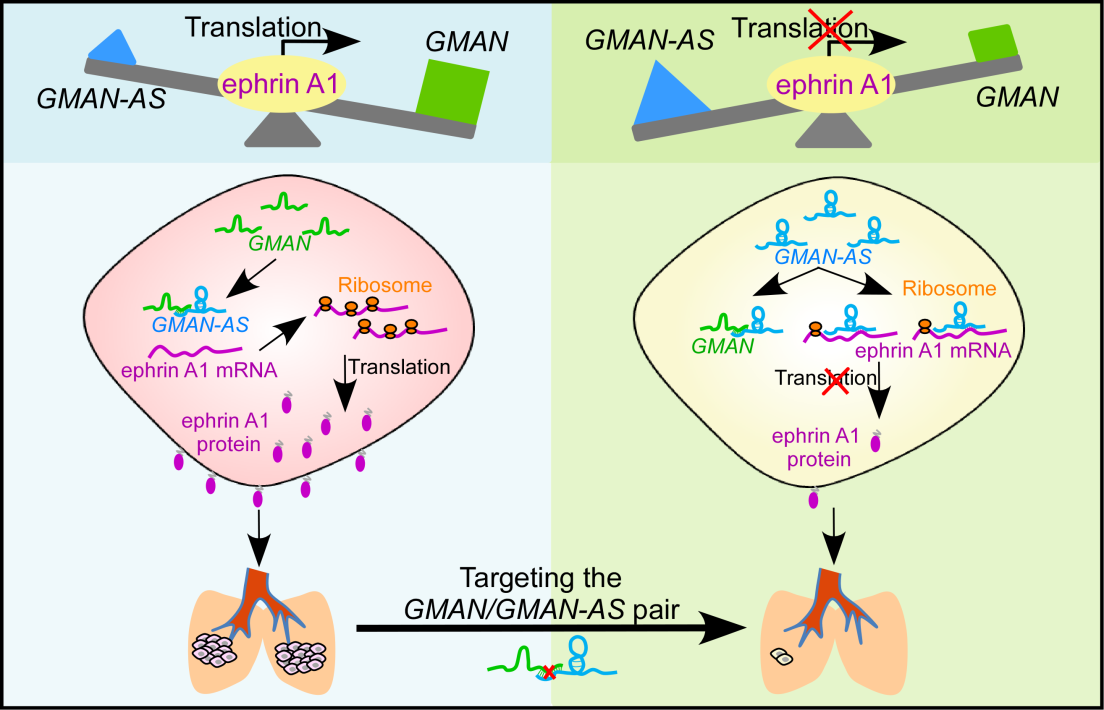
To understand the molecular mechanism of how the sense lncRNA GMAN regulates its overlapping gene ephrin A1, they found another unrecognized RNA antisense to GMAN (GMAN-AS) whose sequence is also partially complementary to the ephrin A1 mRNA. GMAN-AS inhibits ephrin A1 mRNA translation by directly binding to ephrin A1. Further data suggested a triple RNA competition model for translational regulation, whereby the competitive interaction of three RNAs, GMAN, GMAN-AS and ephrin A1 dynamically controls the translation of ephrin A1. In gastric cancer cells with elevated expression of GMAN, most GMAN-AS is buffered by GMAN, allowing ephrin A1 mRNA to form polysomes for efficient production of ephrin A1 protein, which in turn promotes gastric cancer metastasis. In tumor cells with low GMAN expression or elevated level of GMAN-AS, the majority of ephrin A1 mRNAs are occupied by GMAN-AS leading to reduced translation of ephrin A1 and suppression of gastric cancer metastasis. In addition, the GMAN:GMAN-AS ratio also shows a strong correlation with ephrin A1 protein levels in gastric cancer tissues and is significantly associated with poor overall survival of gastric cancer patients.
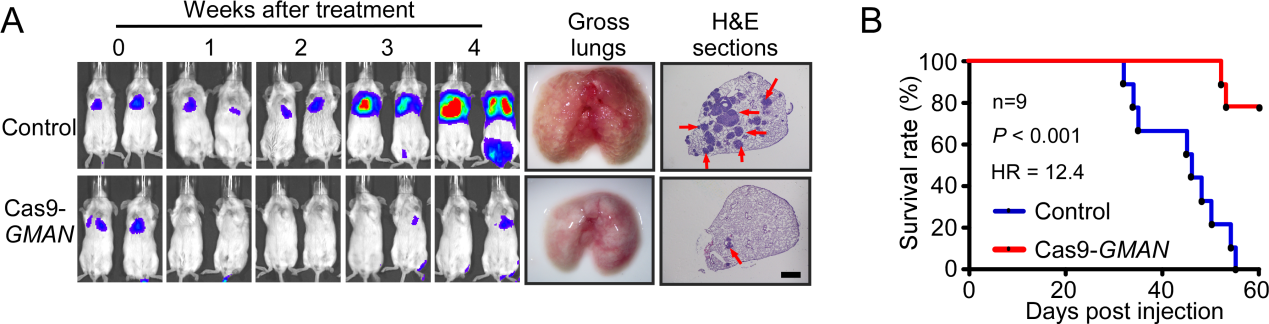
In this manuscript, they provided proof-of-concept animal experiments showing that the therapeutically delivered CRISPR/Cas9 system targeting GMAN robustly suppresses gastric cancer cell metastasis and significantly improved the overall survival in mouse models, highlighting a novel therapeutic approach against gastric cancer metastasis.
Taken all together, they identified two previously undescribed lncRNAs, GMAN and GMAN-AS, as key genes involved gastric cancer metastasis; and uncover a new mechanism of gene expression by which the GMAN/GMAN-AS pair competitively regulates ephrin A1 mRNA translation. This study further provides a rationale for targeting GMAN as a promising therapeutic approach for gastric cancer patients with high risk of metastasis.
This work was mainly done by Wei Zhuo (associate professor) and Yiman Liu (Ph.D. candidate). The first unit of this article is Zhejiang University School of Medicine.
References
1. Torre LA., et al., Global Cancer Statistics, 2012. Ca-a Cancer Journal for Clinicians 2015;65: 87-108.
2. Hayakawa Y., et al., Oesophageal adenocarcinoma and gastric cancer: should we mind the gap? Nature Reviews Cancer 2016;16: 305-318.
3. Chen, W., et al., Cancer statistics in China, 2015. CA: a cancer journal for clinicians, 2016. 66(2):115-132.
4. De Angelis, R., et al., Cancer survival in Europe 1999–2007 by country and age: results of EUROCARE-5—a population-based study. The lancet oncology, 2014. 15(1): 23-34.
5. Bernards, N., et al., No improvement in median survival for patients with metastatic gastric cancer despite increased use of chemotherapy. Annals of oncology, 2013. 24(12): 3056-3060.
6. Djebali, S., et al., Landscape of transcription in human cells. Nature, 2012. 489(7414): 101-108.
7. Gutschner, T. and S. Diederichs, The hallmarks of cancer: a long non-coding RNA point of view. RNA biology, 2012. 9(6): 703-719.
8. Iyer, M.K., et al., The landscape of long noncoding RNAs in the human transcriptome. Nature genetics, 2015. 47(3): 199-208.
9. Chen, L.-L. and G.G. Carmichael, Decoding the function of nuclear long non-coding RNAs. Current opinion in cell biology, 2010. 22(3): 357-364.
10. Laurent, G.S., C. Wahlestedt, and P. Kapranov, The Landscape of long noncoding RNA classification. Trends in Genetics, 2015. 31(5): 239-251.
11. Quinn, J.J. and H.Y. Chang, Unique features of long non-coding RNA biogenesis and function. Nature Reviews Genetics, 2016. 17(1): 47-62.
12. Wang, K.C. and H.Y. Chang, Molecular mechanisms of long noncoding RNAs. Molecular cell, 2011. 43(6): 904-914.
13. Fang, Y. and M.J. Fullwood, Roles, functions, and mechanisms of long non-coding RNAs in cancer. Genomics, proteomics & bioinformatics, 2016. 14(1): 42-54.
14. Mercer, T.R., M.E. Dinger, and J.S. Mattick, Long non-coding RNAs: insights into functions. Nature Reviews Genetics, 2009. 10(3): 155-159.
15. Cesana, M., et al., A long noncoding RNA controls muscle differentiation by functioning as a competing endogenous RNA. Cell, 2011. 147(2): 358-369.

